Automatic selection of calibration files (raw or processed)
This page describes how to automatically associate raw or processed calibrations needed to process the raw science data resutling from an archive query. Science files that need to be processed together, e.g. an infrared jitter sequence, are also grouped together as part of the process. Excerpts from the relevant Night Logs and an xml representation of the calibration cascade are added as ancillary files to the request, as well. The file category provided in the xml file is suitable to be provided as input to the ESO pipelines (for more information on the pipelines, please refer to the relevant User Manuals here). A group of raw science files, calibrations and ancillary files is referred to in this context as a dataset.
The set of calibrations needed to process the raw science files is defined by the calibration plan of that instrument and instrument mode. It is not possible to customize it by, e.g., specifying the type of files needed, nor their number. The calibration plans are distributed as part of the instrument documentation (http://www.eso.org/sci/facilities/paranal/instruments/instrumentName/doc, e.g. http://www.eso.org/sci/facilities/paranal/instruments/hawki/doc).
The time coverage by instrument can be found here. The time coverage is expected to progressively increase as we are constantly striving to extend it as far back in the past as possible. A list of Frequently Asked Questions (FAQs) is also available.
Important notes the current release:
- In order to allow for associations to become available as soon as possible after data acquisition at the observatory (typically within two hours), the associations themselves are at first based on uncertified calibrations. They are, then, replaced with associations based on certified calibrations as soon as the calibrations themselves are checked for quality, typically after two working days (the certification status is recorded by the "certified" flag in the top <association> element in the xml file describing the association). In order to access these improved associations you need to re-run the CalSelector by resubmitting the archive request (see below for the detailed steps).
- In some cases, the validity boundaries for associating calibrations as defined in the calibration plans are formally violated, but the data may be still useful. This is recorded by the "match=extended" flag in the "ASSOCIATION_TREE" file attached to each dataset (see below for a full explanation of the nature and content of this file). In this latter case, care should be applied to verify on a case by case basis the actual applicability of the associated calibrations.
- Under certain circumstances CalSelector returns more science files than actually present in the archive request. This is by design and is intended to provide complete datasets for meaningful processing, e.g. an infrared "on/off-target" sequence where the "off-target" positions may otherwise not be included in the original archive request.
- Under certain circumstances CalSelector does not return an xml file if no association could be established for the input science files (normally it should return an xml file in all cases). The requested input science files are, however, correctly made available for download.
- Under certain circumstances, including a mixture of science and non-science (e.g. acquisition images) files in the same request results in CalSelector behaving erratically. Users are strongly encouraged to include only science files in their query (see below for the details on how to proceed) and let CalSelector associate the other relevant files.
How to obtain the calibration files necessary to calibrate a science raw frame?
A. Via an archive request (old way): The associated raw or processed calibrations historically have been made available to the ESO user community as part of an archive request that the user issues for the related science raw file(s). This method is still available, and below you will find a step-by-step guide for such method.
B. Programmatically: Since Novembre 2019, it is possible to retrieve the associated calibrations programmatically or via VO-aware astronomical tools, using simple http GETs.
C. Via the new Download Portal: Since November 2020 raw data are accessible via the new Download Portal (https://archive.eso.org/downloadportal/[Random Number]) by clicking on “Request this dataset (new service)”. As via an archive request in A., there is both option to get “Associated processed calibrations” or “Associated raw calibrations” by clicking on the corresponding “Run association” button.
A. Obtaining the calibration files at archive request time: A step-by-step guide
1. Point your browser to the archive main page at http://archive.eso.org and follow the link to the raw data query form
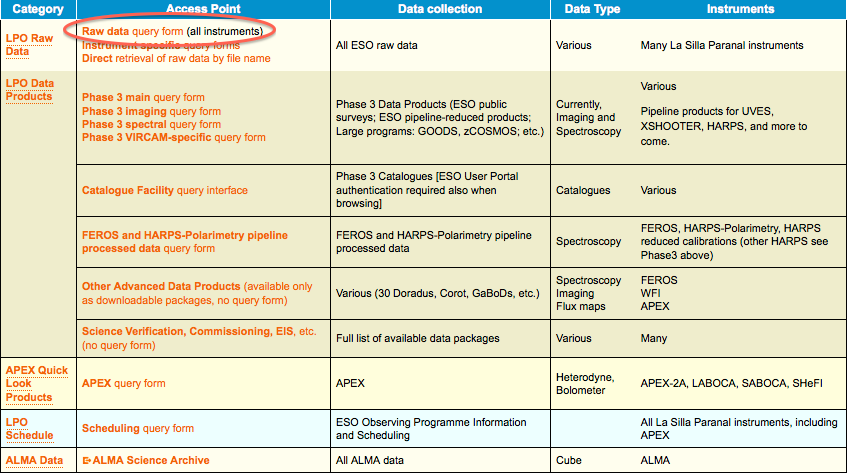
2. In the main query form, if you are interested in a specific programme, e.g. if you are the PI of that programme, just enter its ID and, possibly, a time interval for the nights you are interested in.
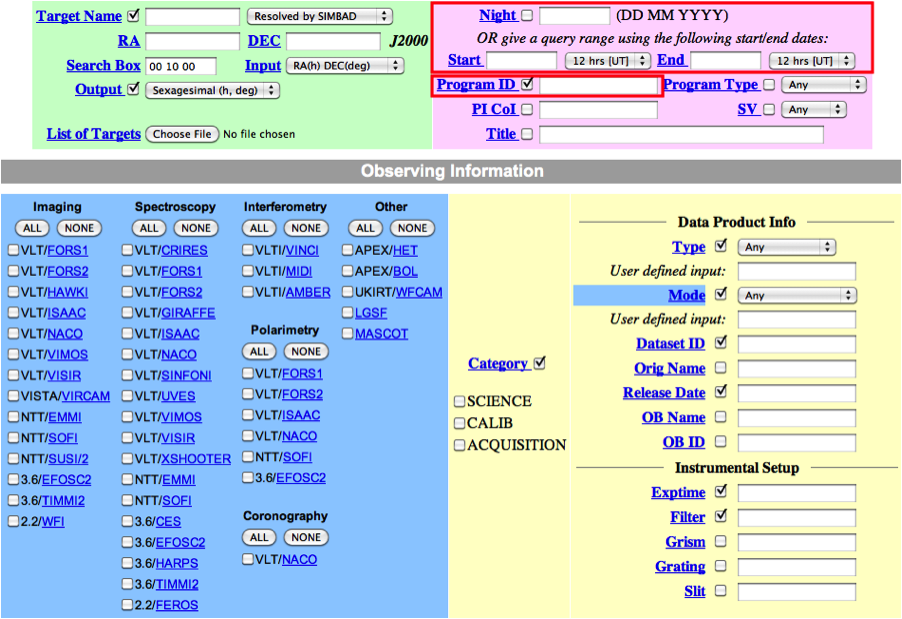
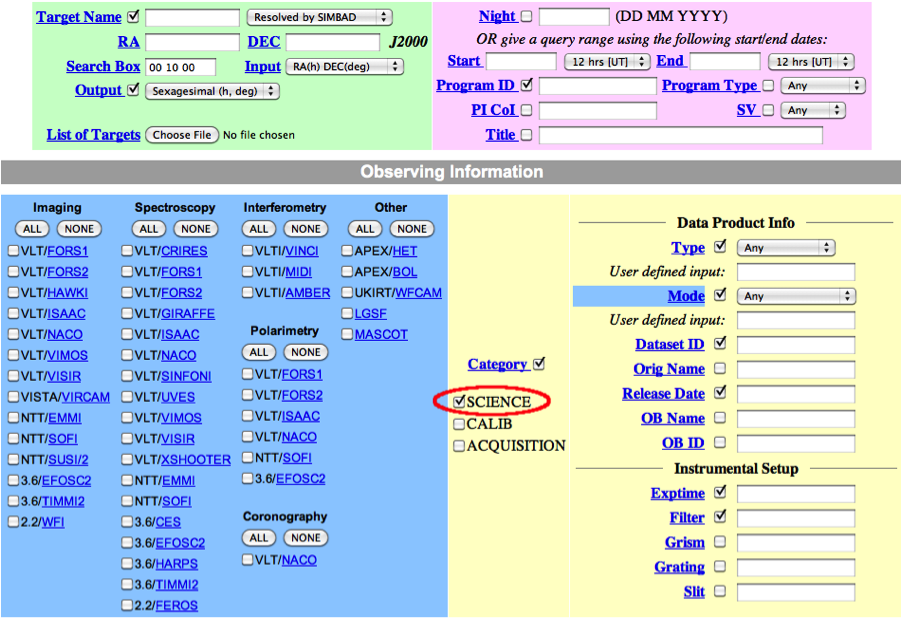
In general, in addition to your other search criteria you should also select Science as Category.
3. In the result page, mark the desired files for download and proceed with selecting the "Request marked datasets" option:

4. In the Download Portal page, then, select which files you want to be delivered: only the selected ones, the selected ones plus the raw calibrations needed to process them, or the selected files plus the processed calibrations (masters) needed to process them. In this latter case, please note that if appropriate processed calibrations are not available, the system will try to associate raw calibrations, instead.
First run the association of the type you want by pressing the corresponding button (raw and/or processed), then toggle the tick box to include/exclude the related files from the download package.

Pressing the "Download Selected" or "Download Request" buttons starts the "Download Manager" applet that allows to choose the destination root directory and the number of cuncurrent download stream and performs the actual download of files. Within this root directory, two subdirectories are created, called "data_with_processed_calibs" and "data_with_raw_calibs", where the datasets with processed calibrations resp. raw calibrations are saved (calibration files common to more then one dataset will be displayed in all relevant datasets, but will only be downloaded once in each of these directories). Alternatively, you can download the data by saving on isk in the intended download directory and executing the provided shell script.
When a dataset is under proprietary period, and the files belongs to you, or otherwise you have gotten permissions as a delegate, you need to Sign In (top right corner) with your ESO User Portal credentials, in order to run the associations.
B. Obtaining the calibration files programmatically
The user scenario in this section describes how to programmatically obtain the calibration files for a given set of science raw frames. Before reading this section, please be aware that the provided eso_download_raw_and_calibs.py python script implements the logic described here.
Using the ESO implementation (tap_obs) of the Tabular Access Protocol (TAP) of the Virtual Observatory, you can perform a direct database query to the table (dbo.raw) that hosts the metadata of the science raw frames. You can learn how to use the TAP interface via a dedicated programmatic page. For example, if you are interested in all the science frames of a particular observing run (e.g., 086.D-0165(A)), the query could be:
| SELECT dp_id FROM dbo.raw WHERE prog_id='086.D-0165(A)' AND dp_cat='SCIENCE' |
where the dp_id is the column name providing the unique raw frame identifier.
You can use the following TAP query for raw data link to learn how to execute a tap_obs query, either interactively from that same page, or most typically via a script (see the Script your access tab in that page), or using a tool like TOPCAT (see the Configure tools tab).
The output of such query (here in text format, but you can also choose FITS or VOTable output formats) is:
dp_id |
Looping through all the output records, you can, for each dp_id, retrieve all the files necessary to calibrate it. How?
First of all, you have to decide whether you want to obtain the raw or the processed calibrations; the calSelector service URL to use is of the form:
| mode | service URL |
| raw calibrations | http://archive.eso.org/datalink/links?ID=ivo://eso.org/ID?the_dp_id&eso_download=calSelector_raw2raw |
| processed (aka master) calibrations |
http://archive.eso.org/datalink/links?ID=ivo://eso.org/ID?the_dp_id&eso_download=calSelector_raw2master |
Try for example: http://archive.eso.org/datalink/links?ID=ivo://eso.org/ID?FORS2.2010-10-23T23:38:41.912&eso_download=calSelector_raw2master
The service URL returns an XML file in a DataLink VOTable format (refer to the DataLink IVOA standard v1.0), a table that contains the list of all the calibration files needed to calibrate the provided input science file, along with their access points (see the access_url field). Sibling science raw frames that need to undergo the same calibration process are also provided within the same table (see the table records with semantics column value set to the string (URI) http://archive.eso.org/rdf/datalink/eso#sibling_raw). It is then possible to loop through all the access_urls, and use them to retrieve the calibration files.
The example above would return many records (see the TABLEDATA part in the snippet below); here are just shown the first two records:
<?xml version="1.0" encoding="utf-8"?> |
The record with the semantics column value set to #this is the record that describes the raw science frame provided in input. Within that record, the column access_url provide the URL to download the science raw frame.
The second, and any other following records with semantics set to #calibration, are all describing the calibration files; loop through them all and use their access_url value to download each of them individually; for example, the command:
curl -JLO "http://archive.eso.org/datalink/datalink/links?ID=ivo://eso.org/ID?FORS2.2010-10-24T09:26:13.380&eso_download=file"
can be used to download that particular BIAS (it is the eso_category field that tells that the file is a bias).
At the end of the table there should be another record, with semantics set to either http://archive.eso.org/rdf/datalink/eso#calSelector_raw2raw or http://archive.eso.org/rdf/datalink/eso#calSelector_raw2master depending on the choice made before, and with the eso_category=ASSOCIATION_TREE; its access_url points to the entire calibration cascade, the XML file that describe the relations between the different science and calibration files (see the ASSOCIATION_TREE below).
When using the service URL, mind the returned http status:
- if the status is 500, there might be some database or similar internal issues. You might want to try again a bit later;
- if the status is 400, something is not correct, please check your syntax;
- If the status is 200, then you got a valid response.
If you receive a valid response, please open such file, and examine the content of the description field of the record identified by the entry with semantics=#this.
That particular description value will tell you 3 things:
- which mode the calSelector could excercise to retrieve the calibration cascade (*);
- It is indeed possible that a request for processed calibrations is downgraded by the service to a request for raw calibrations. This happens when one or more master calibrations are not available. Instead of returning an empty file, the service returns the raw calibrations instead, and sets the mode="raw2raw", which can be found in the description field.
- It is indeed possible that a request for processed calibrations is downgraded by the service to a request for raw calibrations. This happens when one or more master calibrations are not available. Instead of returning an empty file, the service returns the raw calibrations instead, and sets the mode="raw2raw", which can be found in the description field.
- if the calibration association is complete or not (complete="true" or "false");
- whether there was any error while associating the input science frame with its calibration files.
- The description field of the #this record in the example above indeed shows that there was a problem: (complete="false" messages="Missing MASTER_SKY_FLAT_IMG...")
(*) Which mode was executed by calSelector is also reported in the content-disposition HTTP header, which sets the filename to either FORS2.2010-10-24T09:26:13.380_raw2raw.xml or FORS2.2010-10-24T09:26:13.380_raw2master.xml
The structure and content of the file describing the association datasets
As mentioned earlier, an xml representation of the calibration cascade is added to the results of the archive request, and is available via URL in. It is tagged with the category ASSOCIATION_TREE. This section contains a brief explanation of the structure and content of this xml file (an example of an actual file describing an association with raw calibrations is here, while one of an association with master calibrations is here).
Structure: the xml file contains a series of nested <association> elements, where the nesting level reflects the position and role of files in the calibration cascade. Each <association> element, in turn, contains the following elements: <mainFiles> containing, well, the main files for that level of the calibration cascade, <associatedFiles> where the calibrations for the maiFiles are listed and <messages> for additional information on, e.g. the nature of missing files in the case of an incomplete association. The science files are contained in the top <association> element and the cascades ends with the files that do not need any firther calibrations. The names of the actual files are listed in the <file> elements, one file per element.
As for the content, each <association> element has five attributes:
- The category with values suitable to be provided as input to the ESO pipelines.
- The completness flag with possible values "true" or "false", a type flag with possible values "main" "or "auxiliary" (this latter higlighting files that are pertinent to the science files, but are not stictly needed for processing like, e.g. acquisition images for spectroscopic observations).
- The certified flag with possible values "true" or "false", to indicate whether quality-certified calibrations were used to build the calibration cascade provided (typically it takes 2-3 working days to certifiy newly acquired calibrations)
- The type, which can be either "main" or "ancillary".
- The match flag, which indicates whether the associated files are strictly within the boundaries set in the corresponding instrument calibration plan ("match=calib_plan"), or whether these boundaries are formally violated, but the data may still be useful ("match=extended"). In this latter case, care should be applied to verify on a case-by-case basis the actual applicability of the associated calibrations. Static calibrations, such as line lists, configuration tables, etc., carry a flag of "match=N/A".
Note that the first (main) <association> element contains the mode attribute instead of the match flag. The mode is set to "Raw2Raw" if the dataset associates raw calibrations, or to "Raw2Master" if it contains processed calibrations.
The <file> elements, each of which contains one file of the calibration cascade, have two attributes: a category, which is suitable to be provided as input to the ESO pipelines, and the name of the file itself (actually, strictly speaking, that is the root name, without the .fits extension). Please note that the category attribute to the <file> element is, in general, different from the category attribute of the <association> element. The former is a property of the individual file, whereas the latter is a property of that branch of the calibration cascade.
